Abstract
Background: The roles of RNA modifications and RNA-modifying enzymes (writers and erasers) in the regulation of chromatin structure and drug response/resistance in leukemia cells are not well understood. Most published studies have been focused on RNA N6-adenosine methylation (RNA:m6A) and its writers and erasers (Li, S., and Mason, C.E., 2014, Annu Rev Genomics Hum Genet 15, 127). Comparatively little is known about the functions of RNA cytosine methylation (RNA:m5C) and its writers and erasers (Gilbert, et al., 2016, Science 352, 1408). To date, ten RNA:m5C methyltransferases (RCMTs) have been identified, including NSUN1 to NSUN7, three versions (A, B and C) of NSUN5 and DNMT2, but the functions of RNA:m5C and RCMTs remains largely unknown (Schapira, M. 2016. ACS Chem Biol 11, 575). This study aims to determine the impact of RNA:m5C/RCMTs on chromatin structure and 5-azacytidine (5-AZA) response/resistance in myeloid leukemia cells.
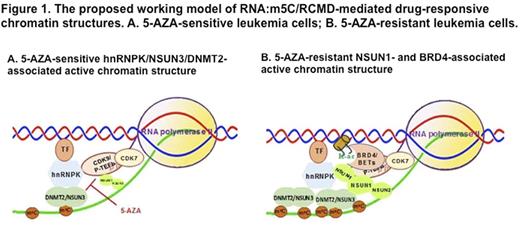
Experiments and Results: Our initial experiments demonstrate a clear relationship between drug resistance and the backbone chemical structures of nucleic acid analogues/drugs and distinct cell lineage-specific drug response patterns in our leukemia cell lines. These results led us to study the roles of RNA:m5C and RCMTs in the regulation of cell lineage-specific chromatin structure and differential drug response in leukemia. Our data demonstrate a marked increase in RNA:m5C and RCMTs as well as their associated active chromatin in our 5-AZA resistant leukemia cell lines, compared to the original 5-AZA sensitive leukemia cell lines. Our RNase-coupled co-immunoprecipitation demonstrates that RCMTs interact with different partners to form functional complexes and distinct chromatin structures, which are essential for the differential drug response and the survival of the leukemia cells. Specifically, NSUN3 and DNMT2 directly interact with a conserved RNA-binding protein hnRNPK to form a 5-AZA-sensitive multi-protein complex. hnRNPK preferentially binds methylated RNA and interacts with multiple proteins, including cell lineage-specific transcription factors GATA1 and SPI1/PU.1 and CDK9/P-TEFb, a kinase phosphorylating the serine 2 (S2) in the C-terminal domain (CTD) of RNA polymerase II (RNA-pol-II). In contrast, NSUN1 and NSUN2 are not associated with hnRNPK directly. Instead, NSUN1 directly binds active (phosphorylated) RNA-pol-II to form active chromatin structure that is completely resistant to 5-AZA. Strikingly, 5-AZA-resistant leukemia cells have a massive increase in NSUN1-and BRD4-associated active RNA-pol-II, compared to the original 5-AZA-sensitive leukemia cells. By employing the newly developed technologies, including 5-ethynyl uridine-clicking chemistry and ligation-mediated rolling circle amplification, we demonstrate that RCMTs recruit active RNA-pol-II to nascent (methylated) RNA to form distinct chromatin structures in the 5-AZA-sensitive vs. 5-AZA-resistant leukemia cells. Knockdown of the expression of these RCMTs causes growth inhibition in both the 5-AZA-sensitive and 5-AZA-resistant leukemia cells and also resensitize the 5-AZA-resistant leukemia cells to a low dose of 5-AZA. Based on our data, we propose the working model of RNA:m5C/RCMT-mediated chromatin structures in 5-AZA-sensitive vs. the 5-AZA-resistant leukemia cells (Figure 1). To test the working model, we applied various CDK and BRD4 inhibitors that target the serine phosphorylation in the CTD of RNA-pol-II to our 5-AZA-sensitive and 5-AZA-resistant-resistant leukemia cell lines. Our data show that these inhibitors synergize with 5-AZA and resensitize the 5-AZA-resistant leukemia cells, leading to growth inhibition in the leukemia cells at very low drug concentrations. By applying these technologies to clinical specimens, we demonstrate the existence of RNA:m5C/RCMT-mediated chromatin structures in clinical MDS/AML cells, supporting their importance and clinical relevance of our working model (Figure 1).
Conclusion: Our data demonstrate that RCMTs interact with RNA-pol-II complex to form distinct chromatin structures at nascent (methylated) RNA, which regulate drug response/resistance in leukemia cells. Such RNA:m5C/RCMT-mediated drug-responsive chromatin structures may lead to the identification of new therapeutic avenues.
Shammo: Alexion Pharmaceuticals, Inc.: Consultancy, Honoraria, Research Funding. Larson: Astellas: Consultancy, Research Funding; Daiichi Sankyo: Consultancy, Research Funding; Novartis: Consultancy, Research Funding; Amgen Inc.: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal